Note on input format
saspt expects input trajectories as a large table of spatiotemporal coordinates. Each coordinate represents the detection of a single fluorescent emitter, and is associated with a trajectory index that has been assigned by a tracking algorithm. An example is shown in the figure below.
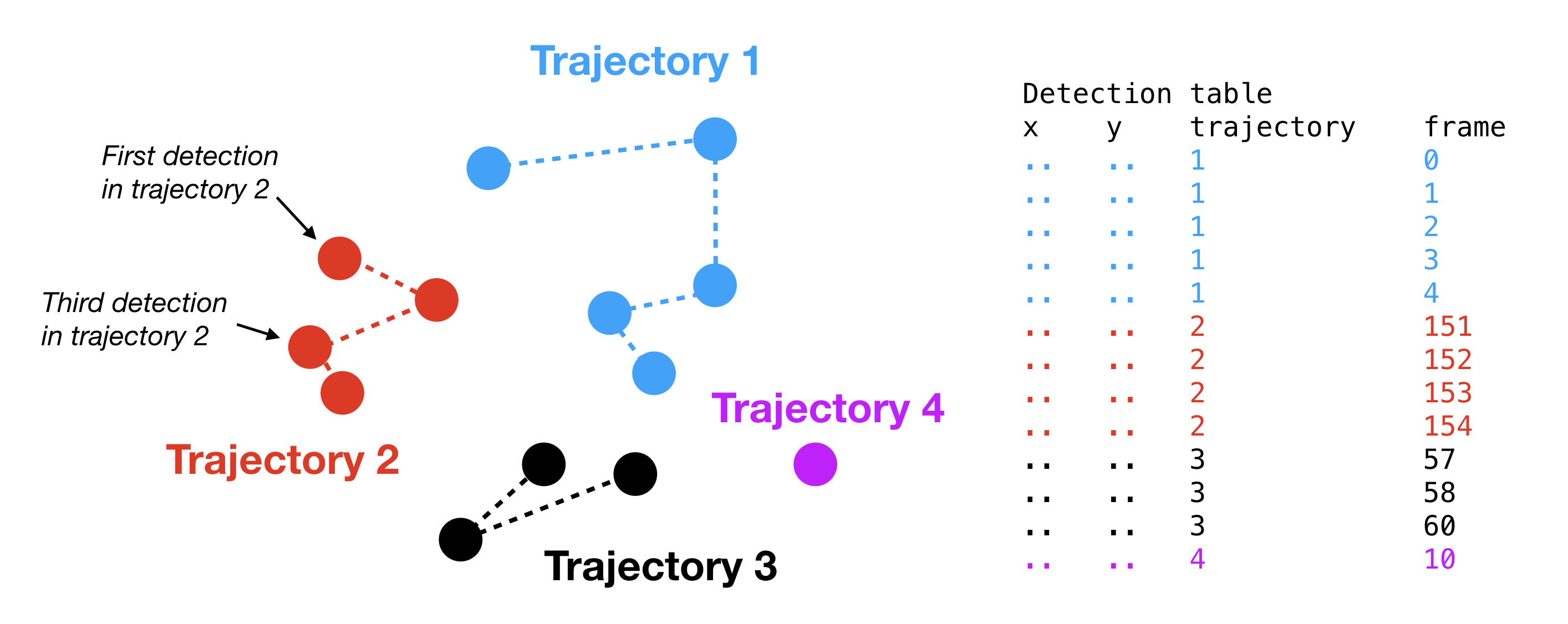
Example of a table of detections. Each dot represents a detection and the dotted lines represent connections (“jumps”) between detections in the same trajectory. Notice that the trajectories may contain “gaps”, or missing frame indices, as in the case of trajectory 3.
The detection table (usually as a CSV) is the format expected by saspt.
This format was chosen for simplicity.
saspt comes with an example of the kind of input it expects:
>>> from saspt import sample_detections
>>> detections = sample_detections()
>>> print(detections)
y x frame trajectory
0 575.730202 84.828673 0 13319
1 538.416604 485.924667 0 1562
2 107.647631 61.892363 0 363
.. ... ... ... ...
493 366.475688 70.559735 297 14458
494 363.350134 67.585339 298 14458
495 360.006572 70.511980 299 14458
[496 rows x 4 columns]
The XY coordinates in pixels.
These can then be used to construct the objects that saspt expects (see
the class hierarchy at API):
>>> from saspt import TrajectoryGroup, StateArrayParameters, StateArray, make_likelihood, RBME
>>> settings = dict(likelihood_type = RBME, pixel_size_um = 0.16, frame_interval = 0.00748)
>>> params = StateArrayParameters(**settings)
>>> trajectories = TrajectoryGroup(detections, **settings)
>>> likelihood = make_likelihood(**settings)
>>> SA = StateArray(trajectories, likelihood, params)
>>> print(params)
StateArrayParameters:
pixel_size_um: 0.16
frame_interval: 0.00748
focal_depth: inf
splitsize: 10
sample_size: 10000
start_frame: 0
max_iter: 200
conc_param: 1.0
>>> print(trajectories)
TrajectoryGroup:
n_detections: 434
n_jumps: 370
n_tracks: 64
>>> print(SA)
StateArray:
likelihood_type : rbme
n_tracks : 64
n_jumps : 370
parameter_names : ('diff_coef', 'loc_error')
shape : (101, 36)
Although this approach is explicit, it is usually easier to use one of the alternative constructors that produce a StateArray directly from a set of detections:
>>> SA = StateArray.from_detections(detections, **settings)
This executes exactly the same steps implicitly:
>>> print(SA)
StateArray:
likelihood_type : rbme
n_tracks : 64
n_jumps : 370
parameter_names : ('diff_coef', 'loc_error')
shape : (101, 36)